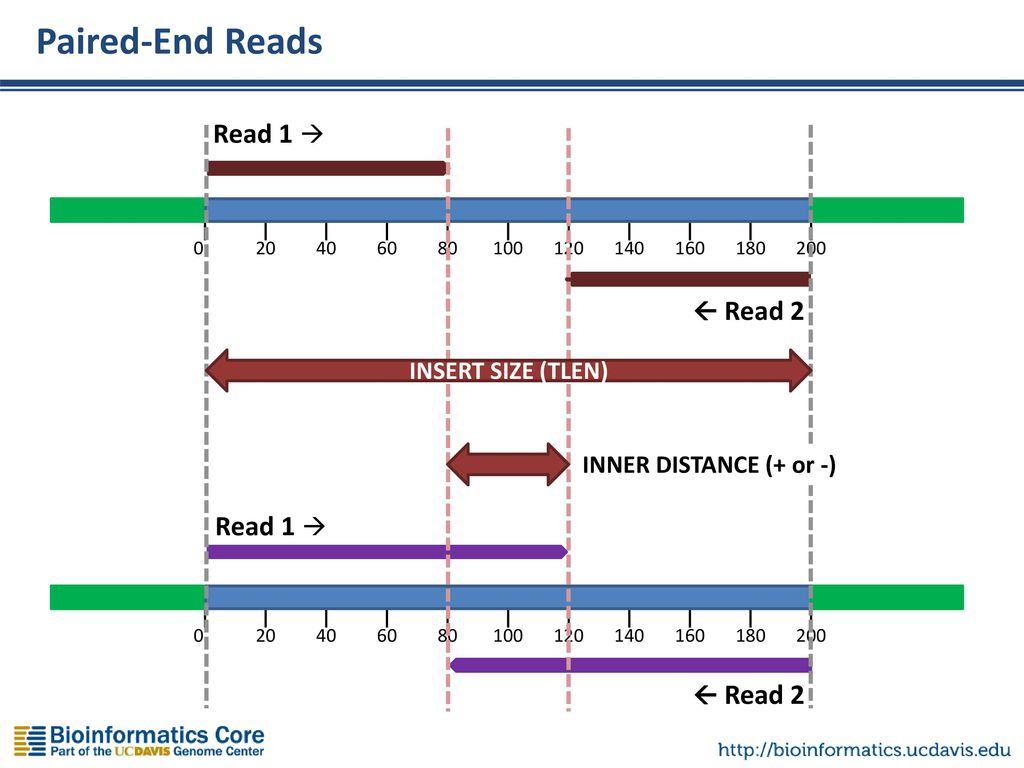
paired end sequencing insert size
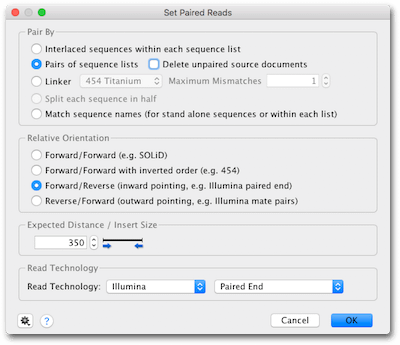
This can be done using the Set Paired Reads. Leftmost coordinate read length - rightmost coordinate.
Introduction To Rna Seq Ppt Download
Read files from paired-end sequencing need to be paired in Geneious before the pairing information can be used in assembly.

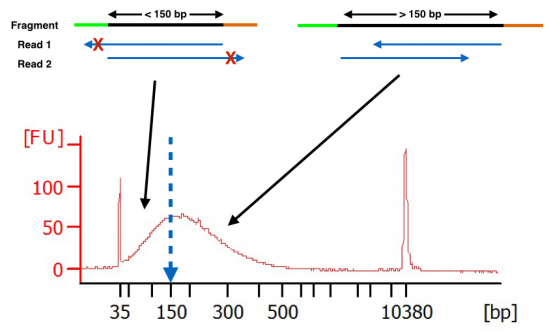
. However the average size of human coding exons is only 160bp. Paired-end RNA sequencing RNA-Seq enables discovery applications such as detecting gene. Thats why you should first.
The size of fragments inserted is 150350 kb. Deep sequencing of transcriptomes allows quantitative and qualitative analysis of many RNA species in a sample with parallel comparison of expression levels splicing. The difference between paired-end and mate-paired is typically that mate-paired is used to indicate a longer insert size compared to paired-end with insert sizes measuring.
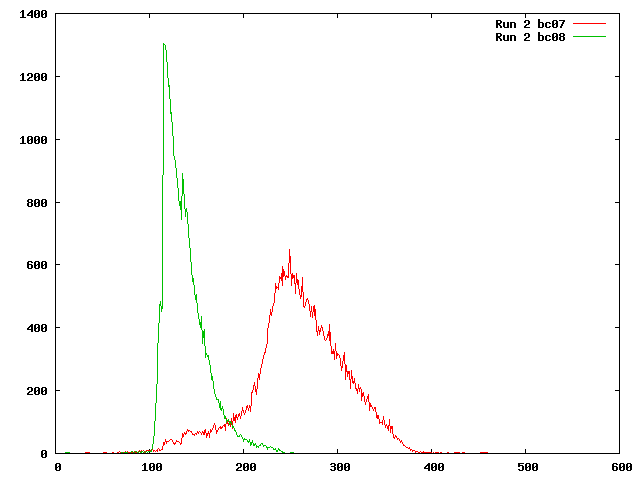
What is insert size paired end. That is it gives the overlap of the two reads. A Sample-wise IS histograms of liquid biopsies LBs.
Including flanking intronic regions of. Relative Orientation - Different sequencing. For instance for a PE150 sequencing run the insert size should be above 300bp.
The variation of insert sizes is often large and the average size difficult to control. Sequencers are supplied DNA samples in fragments of a known length and each end is sequenced generally in a 5 to. Download scientific diagram Paired-end PE sequencing and insert size IS filtering to increase sensitivity.
As different sequencing technology uses different methods of generating reads you should choose the appropriate settings for your data. 07-31-2011 1158 PM. In example 2 the inferred insert size is.
The insert is normally the stretch of sequence between the paired-end adapters so in your case the insert size would be 250 bp 2x75 bp reads 100 bp. The insert is normally the stretch of sequence between the paired-end adapters so in your case the insert size would be 250 bp 275 bp reads 100 bp. To read more about the different parts of a prepared DNA fragment please take a look on the figure in the article about observed sequence lengths in Illumina.
Insert length is the length of the sequence in between a pair of reads. This can result in a proportion of fragments with an insert size of less than the length of a. Despite the benefits of gaining SS-PE data with paired ends of varying distance the standard Illumina protocol allows only non-strand-specific paired-end sequencing with a single insert.
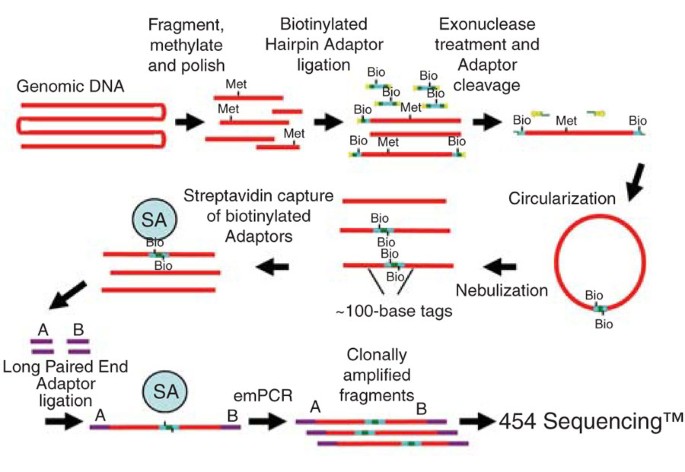
3k Long Tag Paired End Sequencing With The Genome Sequencer Flx System Nature Methods
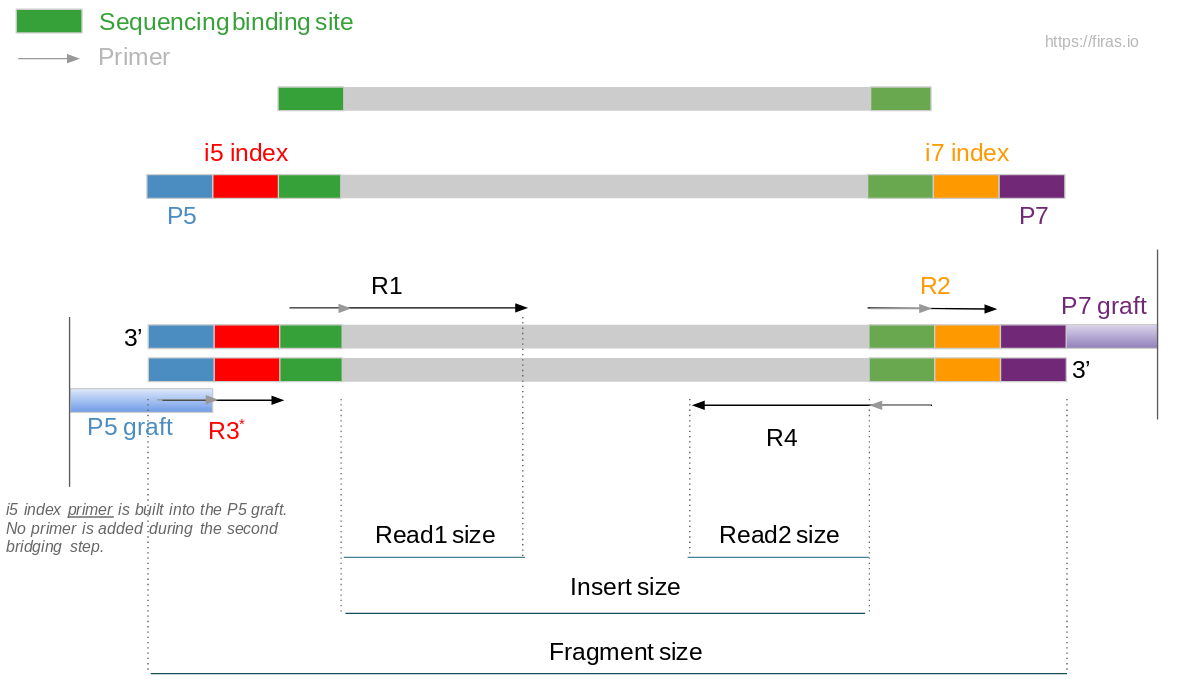
An Overview Of Illumina Multiplexing Firas Sadiyah
Mcb Lecture 9 Sept 23 14 Illumina Library Preparation De Novo Genome Assembly Ppt Download
What Happens When Hardware Advances Faster Than Molecular Protocols Cofactor Genomics

Illumina Sequencing Illumina Sequencing By Synthesis 1010genome Quality Ngs Bioinformatics Data Analysis Services
A Genetic Algorithm For Diploid Genome Reconstruction Using Paired End Sequencing Plos One
De Novo Assembly Tutorial Geneious Prime
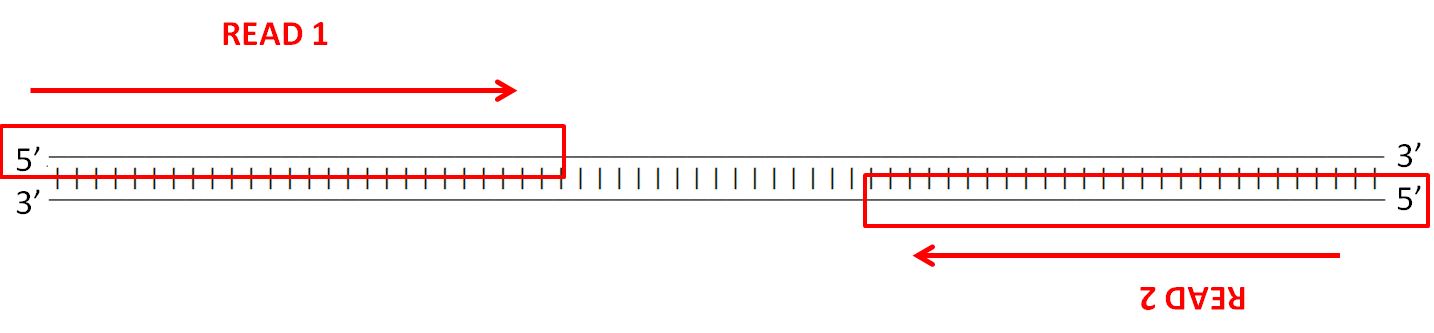
Forward And Reverse Reads In Paired End Sequencing

Illumina Sequencing Illumina Sequencing By Synthesis 1010genome Quality Ngs Bioinformatics Data Analysis Services
Archive Bioinformatic Tools Bwa Banana Slug Genomics
How Sequencing Works Ngs Analysis
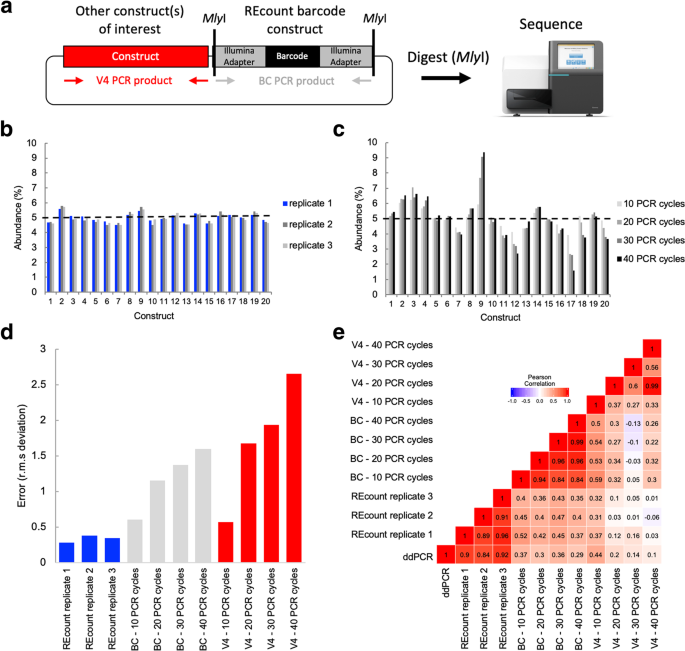
Measuring Sequencer Size Bias Using Recount A Novel Method For Highly Accurate Illumina Sequencing Based Quantification Genome Biology Full Text
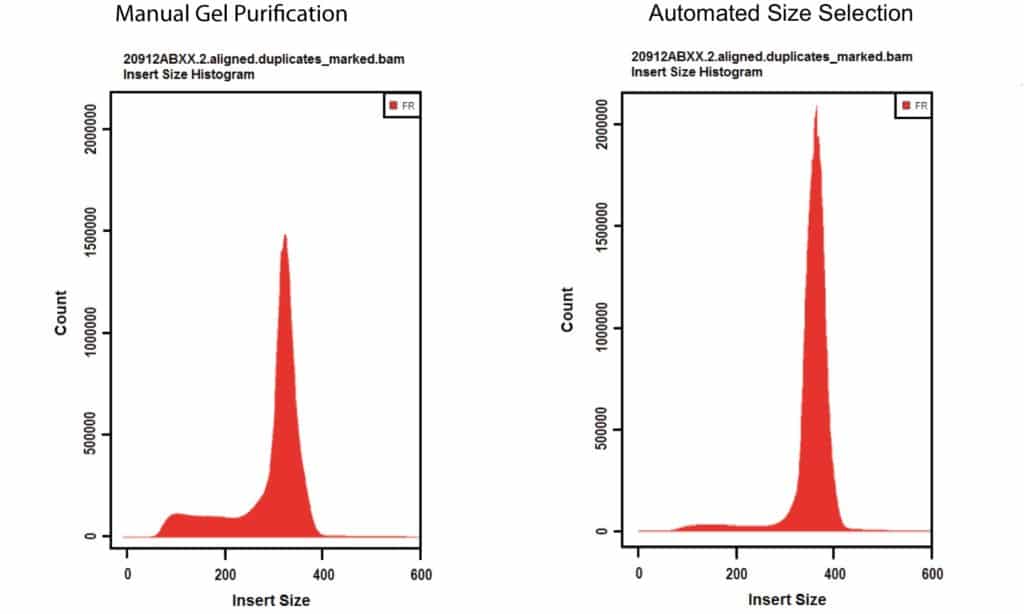
Why Dna Size Selection Matters In Ngs Pipelines
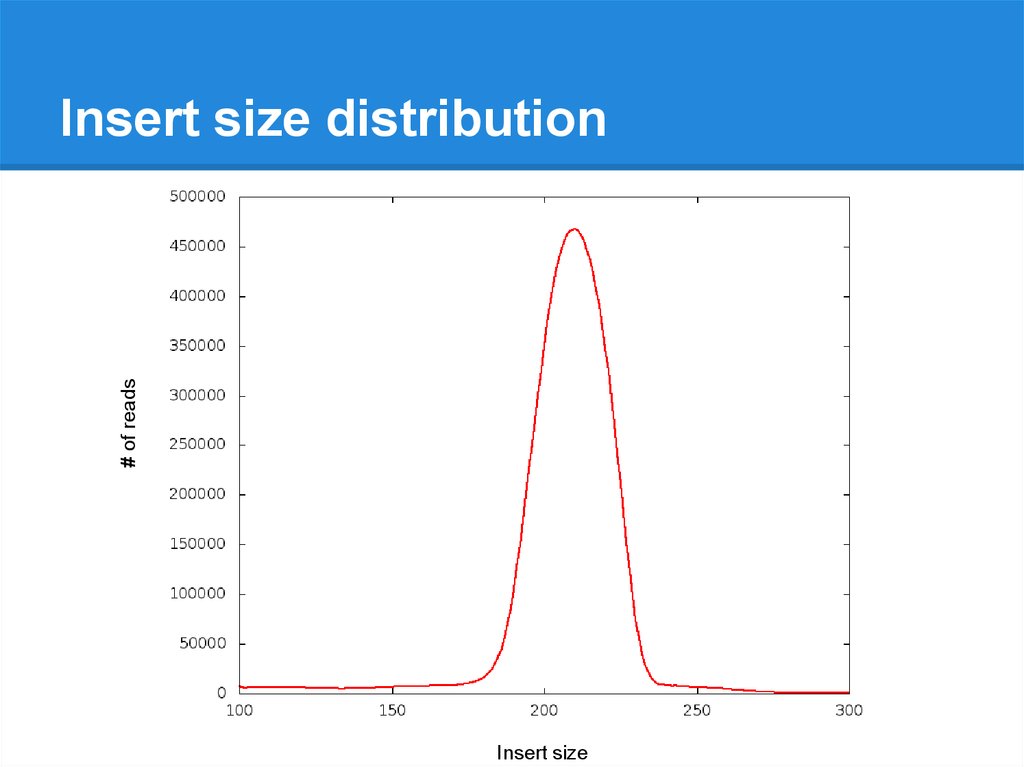
Illumina Data Qc Basic Ngs Tools Prezentaciya Onlajn
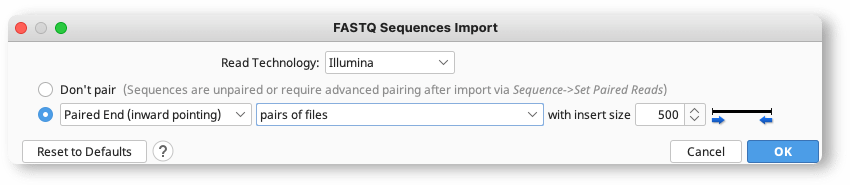
Mapping And Snp Calling Tutorial Geneious Prime
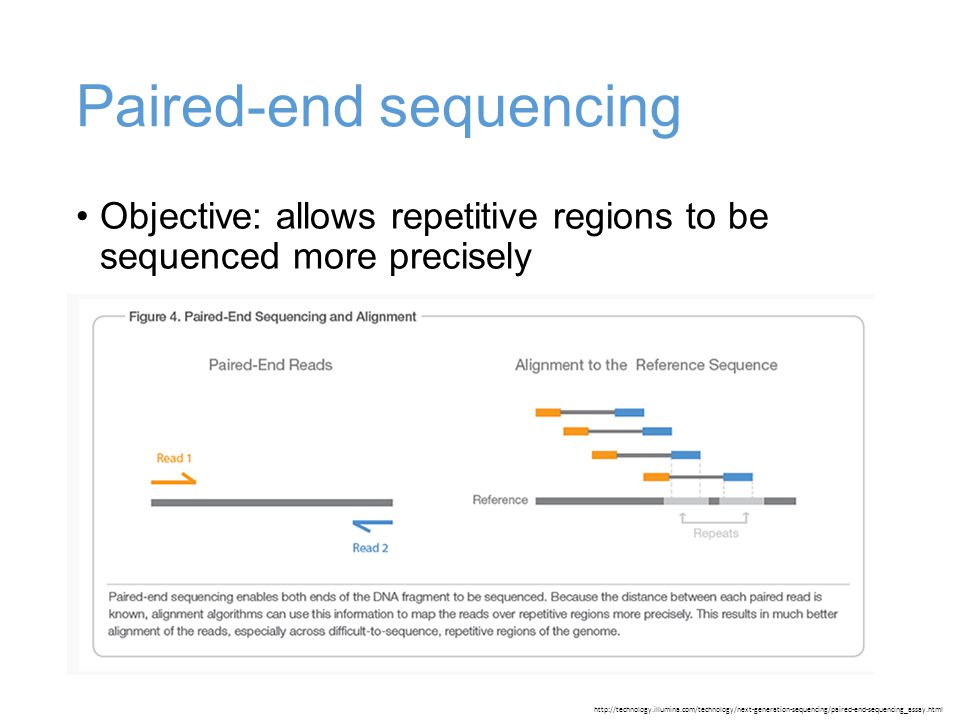
Multiple Insert Size Paired End Sequencing For Deconvolution Of Complex Transcriptomes Rna Biology Vol 9 No 5

What Is Mate Pair Sequencing For

Pdf Simfuse A Novel Fusion Simulator For Rna Sequencing Rna Seq Data Semantic Scholar
